Research activities
Study of karyotypes in the genus Trifolium
Study of karyotypes in the genus Trifolium
- Protocols of chromosome preparations in the genus Trifolium.
- Fluorescence in situ hybridization and study of specific elements (cytogenetic markers) in pratenseand T. medium and their relatives.
- Ancestral state reconstruction of 5S and 26SrDNA sites numbers in the genus Trifolium.
Hybridization of 25S rDNA (red) and probable centromeric repeats (green) of Trifolium pratense to chromosomes of the closely related Trifolium pallidum
Gene variability in the context of biological nitrogen fixation and applications in plant breeding
Gene variability in the context of biological nitrogen fixation and applications in plant breeding
- measurement of nitrogen fixation efficiency between contrasting genotypes of T. pratense by ARA
- targeted sequencing of selected genes involved in nitrogen fixation and nodulation processes in contrasting grass clover genotypes
- identification of polymorphisms (SNPs, SSRs) in selected genes, determination of the degree of association of the identified polymorphisms with the phenotypic trait of high N
- fixation efficiency
use of selected polymorphisms to synthesize a population chip enabling rapid and massive genotyping of plants - verification of the effect of selected polymorphisms on gene expression by real-time PCR
- sequencing of the transcriptome of genotypes contrasting at the N-fixation level, analysis of differential gene expression
The study brought description of phenotypic and genotypic variation in the context of biological nitrogen fixation (BNF) in red clover (Trifolium pratense) and identification of candidate genes associated with BNF efficiency. Plant–rhizobia symbiosis can activate these key genes involved in regulating nodulation associated with biological nitrogen fixation. In summary, knowledge of the genetic variation within BNF candidate genes and the characteristics of genetic variants are beneficial in molecular diagnostics and breeding of red clover.
In the continuity of the paper, functional sample of a population chip was constructed as the substantial innovation of biotechnological methods based on a simultaneous and multiple analysis of the chosen genetic characters associated with the BNF in red clover analysed on a newly developed population chip. The principle of the progressive and environmentally friendly technology is the interaction of the short DNA fragments immobilised on an active matrix with their complementary fluorescent probes designed for detecting a unique gene set. The population chip was designed for effective diagnostics and population screening using genes associated with higher nitrogen fixation; it is a biotechnological tool for improving crop yields based on genomic knowledge on a population level with the positive impact on a content of stable organic matter in the fertile soils.
Technical verification of the functional sample named as a population chip was supported by two registered designs with ID 34500 and ID 34717. The contribution of Masaryk University was in the field of genomic and bioinformatics analyses.
Degree of association of detected polymorphisms in sequenced genes with nitrogen fixation efficiency in T. pratense.
Study of genomes of cultural and wild Trifolium species, de novo sequencing
Study of genomes of cultural and wild Trifolium species, de novo sequencing
- Next-generation sequencing technologies enabled the genome assembly in the pratense genome. The first clover draft genome sequence was obtained in 2014.
- Analysis of repetitive sequences and protein-coding genes in the genome of pratense and gene annotation using bioinformatics tools.
- Analysis of repetitive and coding sequences using cytogenetic methods.
- Development of species specific DNA markers of pratense and T. medium, candidate gene characterization.
Characterization of Trifolium interspecific hybrids
Characterization of Trifolium interspecific hybrids
- Hybrid characterization on the cytogenomic and molecular level.
- Hybrid employment in the breeding.
- Breeding of the variety Pramedi, hybrid between wild species medium and culture T. pratense.
Mezidruhová hybridizace v rámci rodu Trifolium a studium bariér křížitelnosti
Interspecific hybridization in the genus Trifolium and study of interspecific barriers of crossability
- Hybridization of the culture species pratensewith its wild relatives (T. medium, T. sarosiense, T. alpestre) is focused on the introgression of new traits.
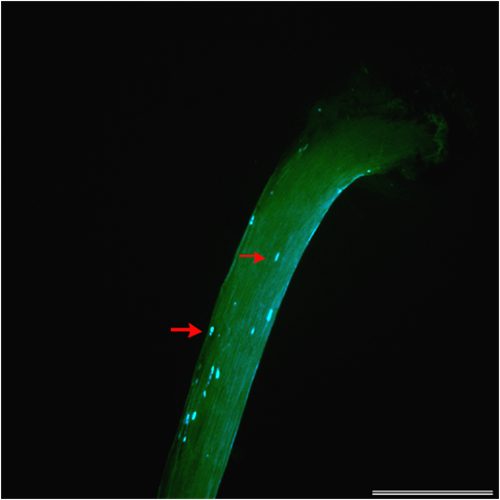
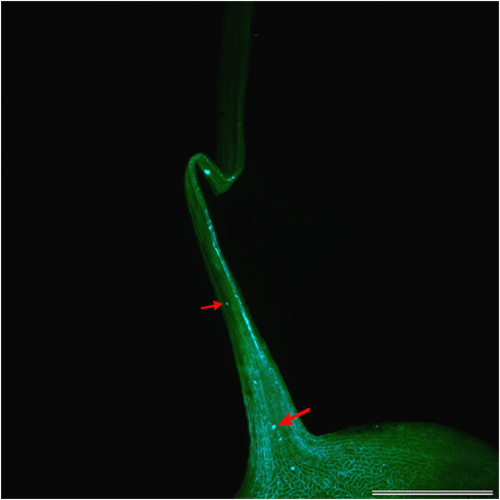
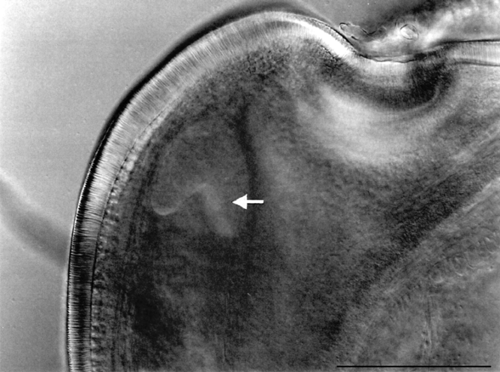
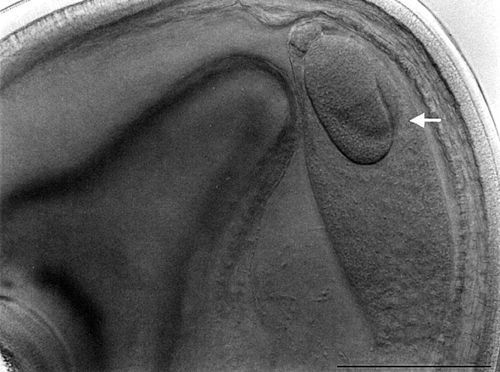
- Study of the pre- and postzygotic barriers crossability after hybridization pratense(4x) x T. medium.
Prezygotic barriers (prevent fertilization): inability of pollen to germinate on the stigma or inability of pollen tubes to grow through the anther
Postzygotic barriers (occurring after fertilisation): defects in endosperm and embryo development
Genetic determination of powdery mildew resistance in Hordeum vulgare
Genetic determination of powdery mildew resistance in Hordeum vulgare
Genetic determination of Hordeum vulgare resistance against powdery mildew caused by Blumeria graminis f. sp. hordei and resistance gene characterization. Identification of new resistance genes in the barley genome using DNA markers and their molecular characterization.
- DNA markers SSR, CAPS, SNP
- Their visualization on the agarose and PAGE gels
- Real-time PCR and Licor detection systems
- TGGE and HRM methods
Publication 1, Publication 2, Publication 3, 4. Publication 4, Publication 5, Publication 6, Publication 7